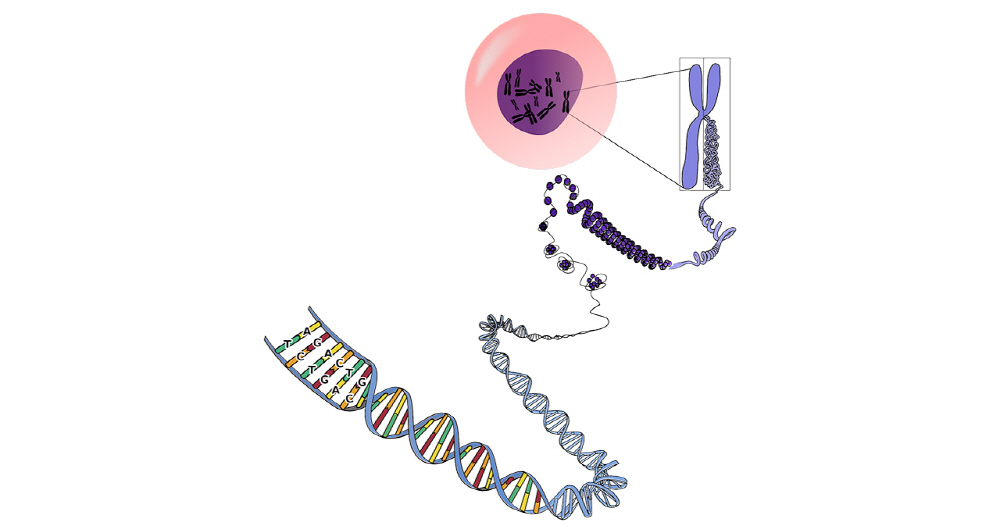
Most cells in the human body have complete copies of DNA that are centered around billions of bases. In addition, each cell of the body can easily access only the necessary part of the DNA embedded in the protein from the outside, and activate genes to become cells with various functions, such as organs, blood, and skin.
Nvidia and Harvard University researchers have developed AtacWorks, an AI toolkit to make it easier to study the DNA accessible parts of the sample data, which are common in early detection of genetic diseases such as cancer, even if there is a lot of noise in the sample data.
The tool runs a screening approach called ATAC-seq (Assay for Transposase-Accessible Chromatin with high-throughput sequencing) on an NVIDIA Tensor Core GPU and runs on a 32-core CPU to find open regions in the genome for healthy and diseased cells. The system completed the genome-wide inference, which would take about 15 hours, in 30 minutes.
In addition, ATAC-seq normally needs to analyze tens of thousands of cells, but if AttackWorks is applied to ATAC-seq, the same quality analysis results can be obtained with only dozens of cells by AI trained in deep learning. For example, the research team was able to identify individual parts of the DNA involved in each production by analyzing only a set of 50 samples of stem cells that make red and white blood cells.
With the effect of reducing the time and cost of genome analysis, AttackWorks can contribute to the identification of cellular lesions and biomarkers that lead to specific diseases. In addition, if genome analysis can be performed even with a small number of cells, research such as identifying DNA differences in rare types of cells will be possible, reducing the cost of data integration, and is expected to bring new possibilities, such as shortening development institutions in the development of new drugs as well as in the diagnostic field do. Related information can be found here.

