
Deep mind has developed AlphaFold, a technology that predicts protein conformation from gene sequence information. If you understand the exact structure of the protein, it is hoped that it will help develop new drugs for Alzheimer’s and Parkinson’s.
Proteins are associated with almost every function a human body can perform, such as turning food into energy, sensing light that shrinks muscles. Protein is a polymer compound in which 20 kinds of L-amino acids are linked in a chain, and how amino acid units called amino acid residues are linked is not recorded in DNA as one-dimensional arrangement information. Therefore, predicting the stereostructure of a protein is a problem of protein folding. Protein folding refers to the process by which a protein, a linear amino acid complex, forms a folded structure that is unique to each protein. In any case, the larger the protein, the more difficult it is to model it.

So far, research has revealed the steric structure of proteins through low temperature electron microscopy, nuclear magnetic resonance, and x-ray crystallography. However, there was a problem that there was trial and error in each structure and cost was high. For this reason, artificial intelligence-based forecasting has attracted attention as an alternative method.
Over the past few years, genetic analysis has evolved dramatically, allowing us to prepare a wealth of genetic data, and Deep Mind has also succeeded in developing alpha- folds based on these data.
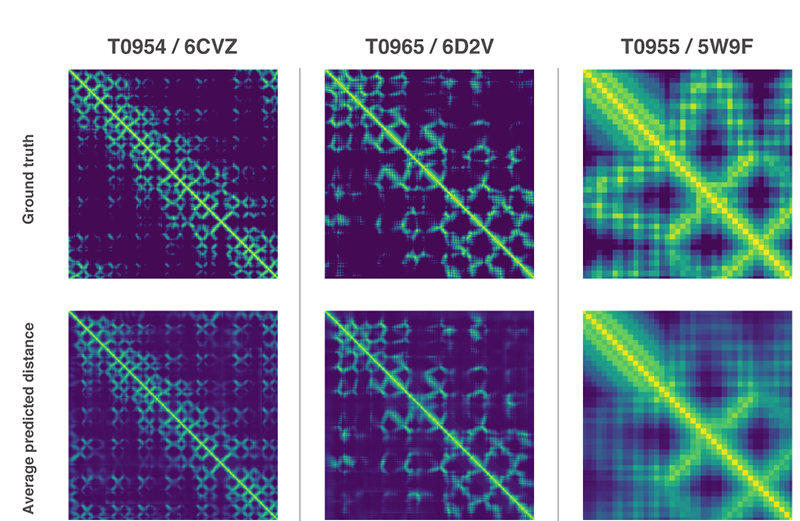
The research team resumed modeling the target structure from the beginning, instead of using the previously solved protein folding problem. As predicted, the team predicted the full protein structure using two methods.
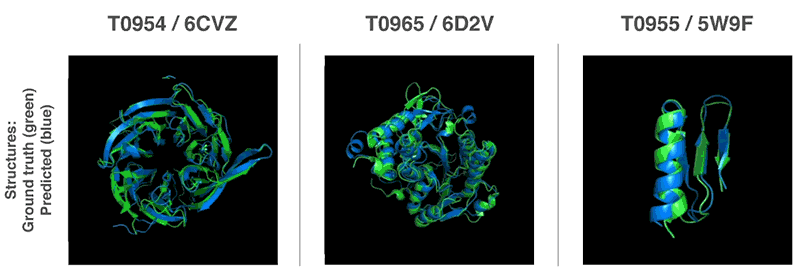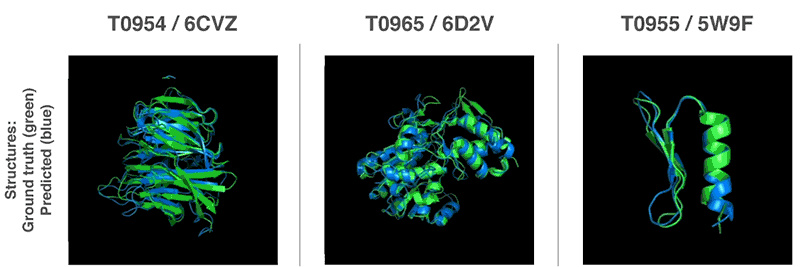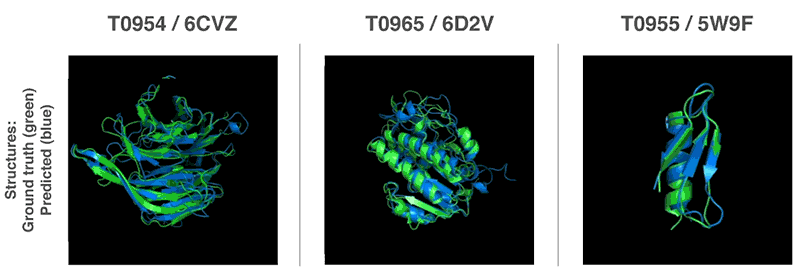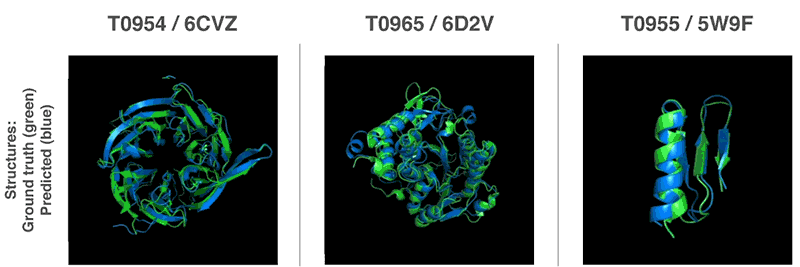
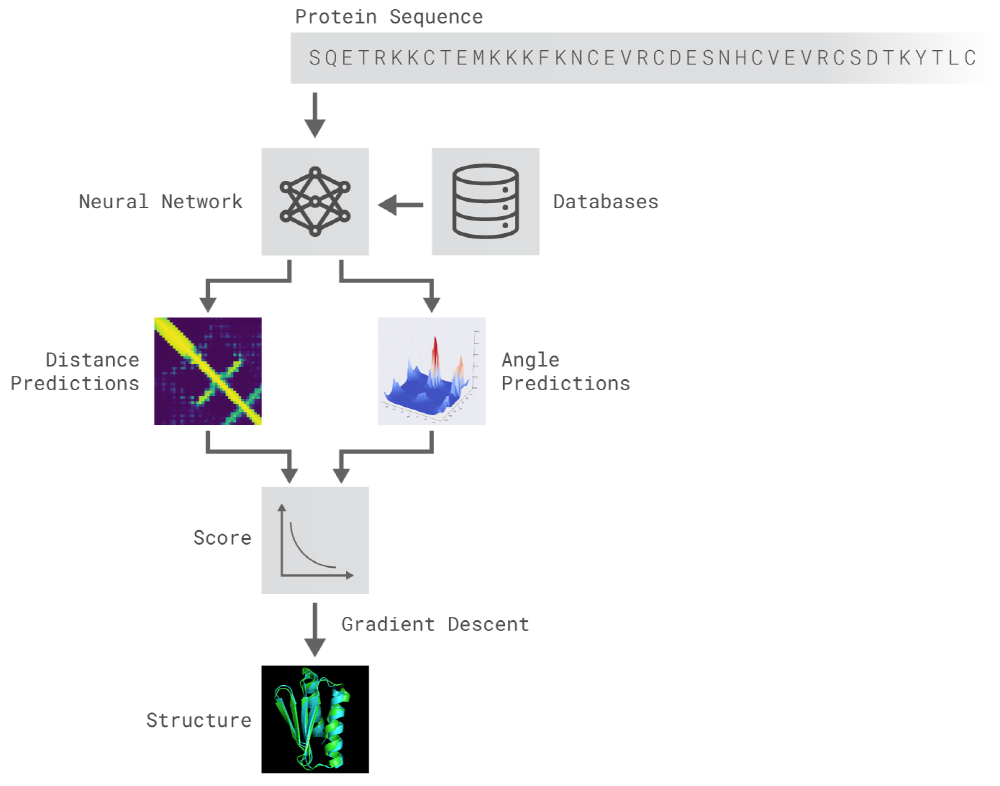
Both methods use neural networks that are trained to predict protein characteristics in gene arrays. The network predicts the properties between the distance of the amino acid pairs and the angle between the chemical bonds connecting them. These two scoring scales how accurate the protein structure can be presented in the two predictions, and that this score can be predicted with high precision using Gradient descent (steepest descent) using machine learning.
As mentioned earlier, Alzheimer’s, Parkinson’s, Huntington’s, and cystic fibrosis are thought to be caused by the wrong fold of the protein. Predicting and understanding the protein structure three-dimensionally will help develop new drugs. Understanding protein folding can also lead to advances in the field of biodegradable enzymes, which may lead to more environmentally friendly decomposition of wastes such as plastics and oils. For more information, please click here .


















Add comment